
Large-Scale Identification, Mapping, and Genotyping of Single-Nucleotide Polymorphisms in the Human Genome
David G. Wang, Jian-Bing Fan, Chia-Jen Siao, Anthony Berno, Peter Young,Ron Sapolsky, Ghassan Ghandour, Nancy Perkins, Ellen Winchester, Jessica Spencer, Leonid Kruglyak, Lincoln Stein, Linda Hsie, Thodoros Topaloglou, Earl Hubbell, Elizabeth Robinson, Michael Mittmann, Macdonald S. Morris, Naiping Shen, Dan Kilburn, John Rioux, Chad Nusbaum, Steve Rozen, Thomas J. Hudson, Robert Lipshutz, Mark Chee, Eric S. Lander
Individual spots on chip put these DNAs
Row 1 ACGTACGTACGTAACGTACGTACGT
Row 2 ACGTACGTACGTCACGTACGTACGT
Row 3 ACGTACGTACGTGACGTACGTACGT
Row 4 ACGTACGTACGTTACGTACGTACGT
Remember A pairs with T and C with G.
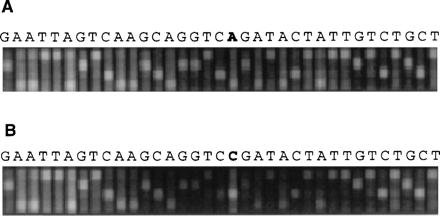
Fig. 1. SNP screening on chips. (A) Small portion of a VDA for an STS hybridized with the expected target sequence. Chip features in each column are complementary to successive overlapping 25-nucleotide oligomer subsequences, with the central base substituted by A, C, G, or T in the four rows. Variations from the expected sequence can usually be detected by examination of the most intense signal in each column. (B) The same VDA was hybridized with sequence containing an SNP (AC) at position 19. The hybridization signal is now stronger at an alternative base at this position. It is also weaker at the surrounding positions (for example, positions 12 to 18 and 20 to 26), because probes at these positions are designed to be complementary to the A allele at the SNP and mismatch with the C allele.
Text iGenetics by Peter J. Russell
This web site is provided for instruction in Botany and Zoology 342
by Kenneth G. Wilson,
Professor of Botany
Miami University
wilsonkg@muohio.edu