|
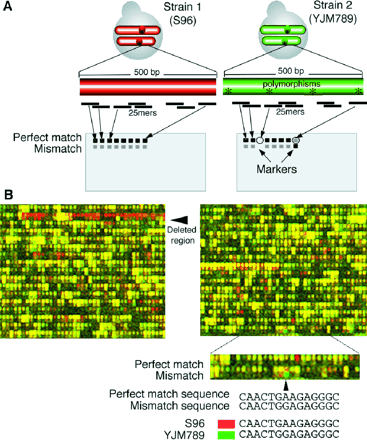
Direct Allelic Variation Scanning of the Yeast Genome Elizabeth A. Winzeler,* Dan R. Richards, Andrew R. Conway, Alan L. Goldstein, Sue Kalman, Michael J. McCullough, John H. McCusker, David A. Stevens, Lissa Wodicka, David J. Lockhart, Ronald W. Davis As more genomes are sequenced, the identification and characterization of the causes of heritable variation within a species will be increasingly important. It is demonstrated that allelic variation in any two isolates of a species can be scanned, mapped, and scored directly and efficiently without allele-specific polymerase chain reaction, without creating new strains or constructs, and without knowing the specific nature of the variation. A total of 3714 biallelic markers, spaced about every 3.5 kilobases, were identified by analyzing the patterns obtained when total genomic DNA from two different strains of yeast was hybridized to high-density oligonucleotide arrays. The markers were then used to simultaneously map a multidrug-resistance locus and four other loci with high resolution (11 to 64 kilobases). E. A.
Winzeler, D. R. Richards, A. R. Conway, S. Kalman, R. W. Davis,
Department of Biochemistry, Stanford University School of Medicine,
Stanford, CA 94305-5307, USA. A. L. Goldstein and J. H. McCusker,
Department of Microbiology, 3020, Duke University Medical Center,
Durham, NC 27710, USA. M. J. McCullough and D. A. Stevens,
Department of Medicine, Stanford University School of Medicine,
Stanford, CA 94305, USA. L. Wodicka and D. J. Lockhart, Affymetrix,
3380 Central Expressway, Santa Clara, CA 95051, USA.
|