Hongyun Wang's Research on Molecular Motors
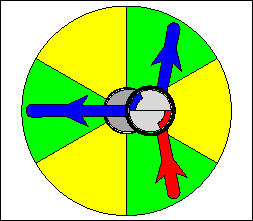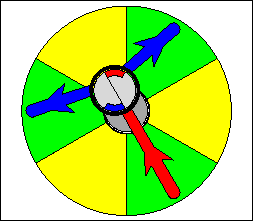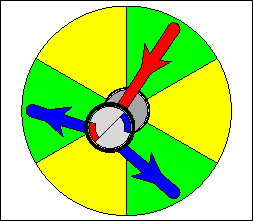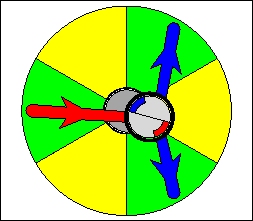
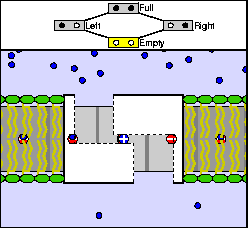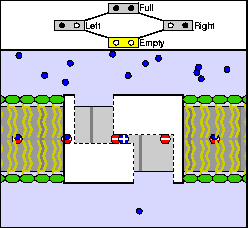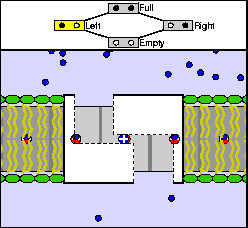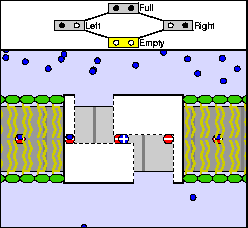
The above are cartoon animations of F1 motor and Fo motor of the ATP
synthase.
Click here for more movies of the ATP synthase.
Hongyun Wang's research in biophysics and molecular modeling is
to investigate the mechanism by which chemical energy is converted
into mechanical work in biological systems. Cells store chemical
energy in two forms: as transmembrane electrochemical gradients
and in chemical bonds, particularly the gamma phosphate bond in
adenosine triphosphate (ATP). ATP is the most important chemical
energy source in all living cells. In mitochondria, bacteria, and
chloroplasts the free energy stored in transmembrane electrochemical
gradients is used to synthesize ATP from ADP and phosphate via the
membrane-bound enzyme ATP synthase. ATP synthase can also reverse
itself and hydrolyze ATP to pump ions against an electrochemical
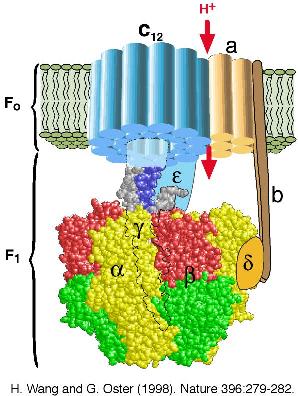
gradient. ATP synthase consists of two portions: a membrane-spanning
portion, Fo, comprising the ion channel, and a soluble portion, F1,
containing three catalytic sites (see the Figure below). Both Fo and F1 are
reversible rotary motors --- perhaps the smallest motors known to science.
Fo uses the transmembrane electrochemical gradient to generate a
rotary torque to drive ATP synthesis in F1 or, when driven backwards
by the torque generated in F1, to pump ions uphill against their
transmembrane electrochemical gradient. F1 generates a rotary torque
by hydrolyzing ATP at its three catalytic sites or, when turned
backwards by the torque generated in Fo, as a synthesizer of ATP.
 |
Click on the picture or here
to view high resolution image.
|
Working with George Oster's group at UC Berkeley, Hongyun
Wang constructed separate models for the Fo and F1 portions of ATP
synthase which explained how the energy is transduced from the
transmembrane electrochemical gradient to a mechanical torque in Fo
(see Refs [1] and [3] ), and how the energy is transduced from ATP to a
mechanical torque in F1 (see Refs [2], [4], [5] and [6]). These are the first
quantitative models of this enzyme, and are in good agreement with the
existing experiments.
In carrying out this research Hongyun Wang developed a computer program
to deduce the kinematic motions of the F1 motor from
John Walker's
crystal structure of F1. Some of the results and animation movies were
included in the interactive CD of the book "Essential Cell Biology"
by Alberts, et al. All animation movies can be accessed at
http://www.cnr.berkeley.edu/~hongwang/Project/ATP_synthase.
Hongyun Wang's recent research is focused on how the free energy
liberated in the ATP hydrolysis cycle is converted efficiently to
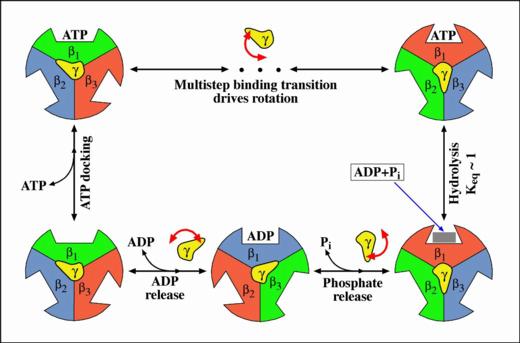
generate a force. The binding zipper model was proposed to explain
the high efficiency of the F1 motor (see Refs [5] and [6]):
- The binding zipper model
In the hydrolysis direction, an ATP in solution first diffuses to
the catalytic site and is weakly bound (ATP docking). The rate of
this step is affected by the ATP concentration in solution. The
weakly bound ATP may dissociate from the catalytic site, returning
to the solution. Occasionally, it proceeds from weak binding to tight
binding (the binding transition). During the binding transition, the
bonds between the ATP and the catalytic site form sequentially, ATP
binding affinity increases gradually, and
each bond formation drives a small conformational change.
In this way, the binding free energy is used efficiently to generate
a nearly constant force during the multi-step ATP binding transition.
ATP concentration in solution does not affect the binding transition,
but only how often ATP attempts docking to the catalytic site.
After the binding transition, the ATP is in chemical equilibrium with
ADP and Pi. The transition ATP <--> ADP + Pi weakens the ATP binding
and distributes it over ADP and Pi so that the hydrolysis products
can be released and the cycle repeated.
Click on the picture or here
to view high resolution image.
This project is supported by NSF.
| [1] |
T. Elston, H. Wang and G. Oster (1998). Energy transduction in ATP synthase.
Nature 391:510-513.
|
| [2] |
H. Wang and G. Oster (1998). Energy transduction in the F1 motor
of ATP synthase.
Nature 396:279-282.
|
| [3] |
P. Dimroth, H. Wang, M. Grabe and G. Oster (1999). Energy transduction
in the sodium F-ATPase of Propionigenium modestum.
Proc. Natl. Acad. Sci. USA 96:4924-4929.
|
| [4] |
G. Oster and H. Wang (1999). ATP synthase: two motors, two fuels.
Structure 7:R67-R72.
|
| [5] |
G. Oster and H. Wang (2000) Reverse engineering a protein: The
mechanochemistry of ATP synthase.
Biochimica et Biophysica Acta 1458:482-510.
|
| [6] |
G. Oster and H. Wang (2000) Why is the efficiency of the F1 ATPase so high?
Bioenerg. Biomembr. 32:459-469.
|





